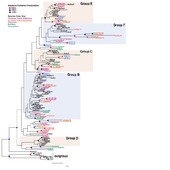
| dc.description.abstract | Background: The Sox genes, a family of transcription factors characterized by the presence of a high mobility group (HMG) box domain, are among the central groups of developmental regulators in the animal kingdom. They are indispensable in progenitor cell fate determination, and various Sox family members are involved in managing the critical balance between stem cells and differentiating cells. There are 20 mammalian Sox genes that are divided into five major groups (B, C, D, E, and F). True Sox genes have been identified in all animal lineages but not outside Metazoa, indicating that this gene family arose at the origin of the animals. Whole-genome sequencing of the lobate ctenophore Mnemiopsis leidyi allowed us to examine the full complement and expression of the Sox gene family in this early-branching animal lineage. Results: Our phylogenetic analyses of the Sox gene family were generally in agreement with previous studies and placed five of the six Mnemiopsis Sox genes into one of the major Sox groups: SoxB (MleSox1), SoxC (MleSox2), SoxE (MleSox3, MleSox4), and SoxF (MleSox5), with one unclassified gene (MleSox6). We investigated the expression of five out of six Mnemiopsis Sox genes during early development. Expression patterns determined through in situ hybridization generally revealed spatially restricted Sox expression patterns in somatic cells within zones of cell proliferation, as determined by EdU staining. These zones were located in the apical sense organ, upper tentacle bulbs, and developing comb rows in Mnemiopsis, and coincide with similar zones identified in the cydippid ctenophore Pleurobrachia. Conclusions: Our results are consistent with the established role of multiple Sox genes in the maintenance of stem cell pools. Both similarities and differences in juvenile cydippid stage expression patterns between Mnemiopsis Sox genes and their orthologs from Pleurobrachia highlight the importance of using multiple species to characterize the evolution of development within a given phylum. In light of recent phylogenetic evidence that Ctenophora is the earliest-branching animal lineage, our results are consistent with the hypothesis that the ancient primary function of Sox family genes was to regulate the maintenance of stem cells and function in cell fate determination. | en_US |